15 Robust Regression
Prerequisites
library("rstan")
library("tidyverse")
library("rstanarm")
library("bayz")
library("loo")
library("jrnold.bayes.notes")
library("recipes")15.1 Wide Tailed Distributions
Like OLS, Bayesian linear regression with normally distributed errors is sensitive to outliers. This is because the normal distribution has narrow tail probabilities, with approximately 99.8% of the probability within three standard deviations.
Robust regression refers to regression methods which are less sensitive to outliers. Bayesian robust regression uses distributions with wider tails than the normal instead of the normal. This plots the normal, Double Exponential (Laplace), and Student-t (\(df = 4\)) distributions all with mean 0 and scale 1, and the surprise (\(- log(p)\)) at each point. Both the Student-\(t\) and Double Exponential distributions have surprise values well below the normal in the ranges (-6, 6).11 This means that outliers will have less of an affect on the log-posterior of models using these distributions. The regression line would need to move less incorporate those observations since the error distribution will not consider them as unusual.
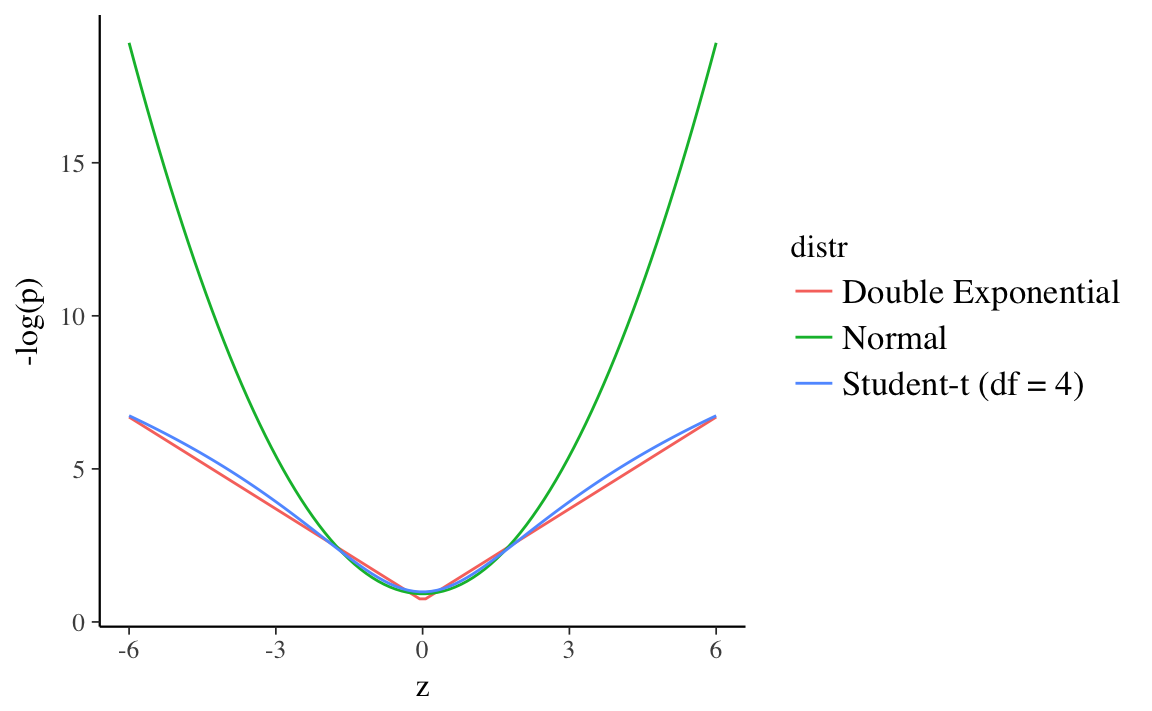
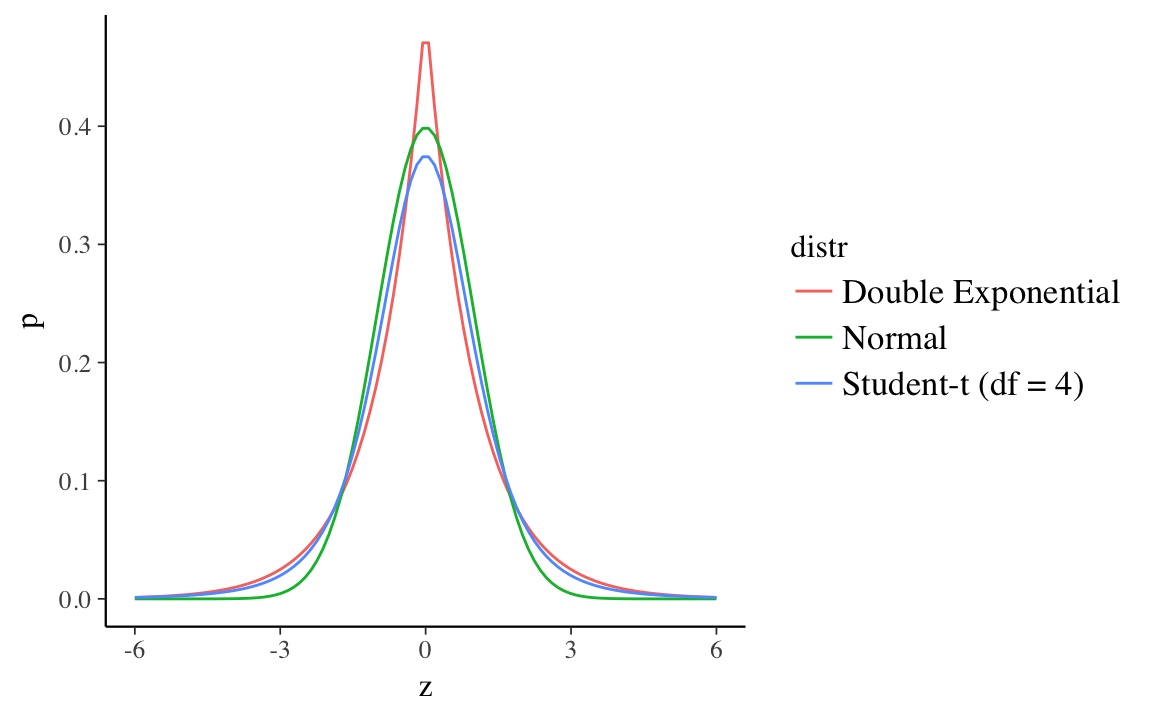
15.2 Student-t distribution
The most commonly used Bayesian model for robust regression is a linear regression with independent Student-\(t\) errors (Geweke 1993; A. Gelman, Carlin, et al. 2013, Ch. 17): \[ y_i \sim \dt\left(\nu, \mu_i, \sigma \right) \] where \(\nu \in \R^{+}\) is a degrees of freedom parameter, \(\mu_i \in \R\) are observation specific locations often modeled with a regression, and and \(\sigma \in R^{+}\) is a the scale parameter.
Note that as \(\nu \to \infty\), this model approaches an independent normal model, since the Student-t distribution asymptotically approaches the normal distribution as the degrees of freedom increases. For the value of \(\nu\), either a low degrees of freedom \(\nu \in (4, 6)\) can be used, or it can be given a prior distribution. For the Student-t distribution, the existence of various moments depends on the value of \(\nu\): the mean exists for \(\nu > 1\), variance for \(\nu > 2\), and kurtosis for \(\nu > 3\). As such, it is often useful to restrict the support of \(\nu\) to at least 1 or 2 (or even higher) ensure the existence of a mean or variance.
A reasonable prior distribution for the degrees of freedom parameter is a Gamma
distribution with shape parameter 2, and an inverse-scale (rate) parameter of 0.1 (Juárez and Steel 2010,@Stan-prior-choices),
\[
\nu \sim \dgamma(2, 0.1) .
\]
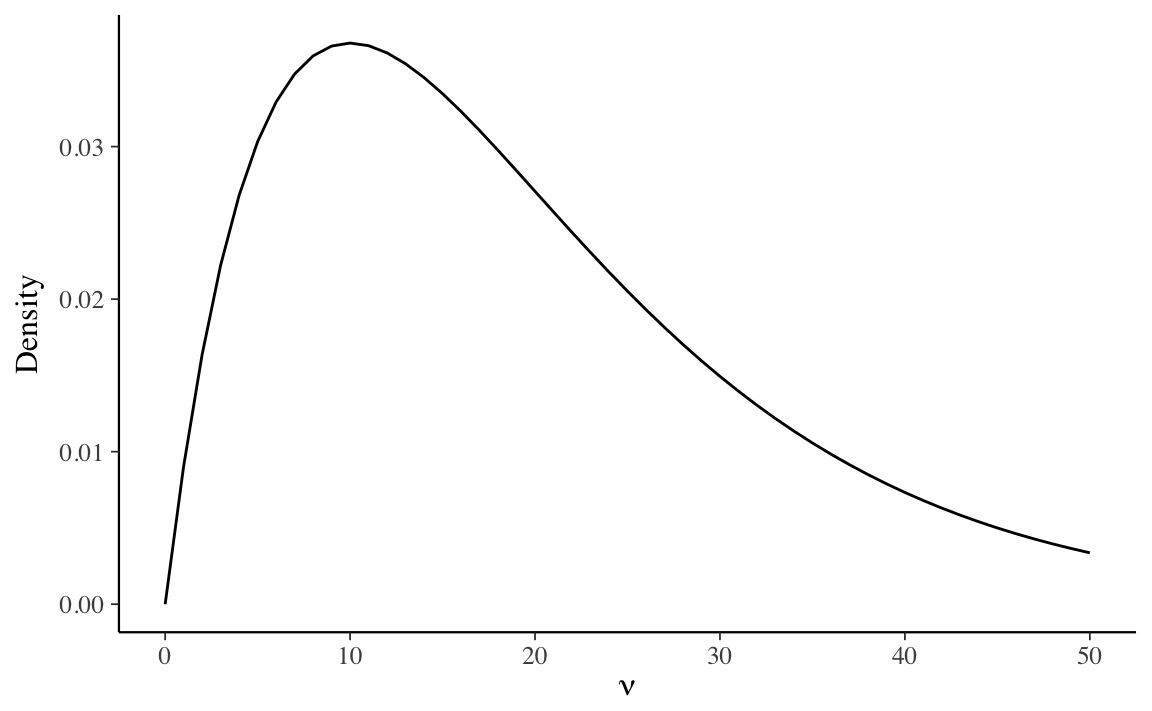 This density places the majority of the prior mass for values \(\nu < 50\), in which
the Student-\(t\) distribution is substantively different from the Normal distribution,
and also allows for all prior moments to exist.
This density places the majority of the prior mass for values \(\nu < 50\), in which
the Student-\(t\) distribution is substantively different from the Normal distribution,
and also allows for all prior moments to exist.
The Stan model that estimates this is lm_student_t_1.stan:
// lm_student_t_1.stan
// Linear Model with Student-t Errors
data {
// number of observations
int N;
// response
vector[N] y;
// number of columns in the design matrix X
int K;
// design matrix X
// should not include an intercept
matrix [N, K] X;
// priors on alpha
real scale_alpha;
vector[K] scale_beta;
real loc_sigma;
// keep responses
int use_y_rep;
int use_log_lik;
}
parameters {
// regression coefficient vector
real alpha;
vector[K] beta;
real sigma;
// degrees of freedom;
// limit df = 2 so that there is a finite variance
real nu;
}
transformed parameters {
vector[N] mu;
mu = alpha + X * beta;
}
model {
// priors
alpha ~ normal(0.0, scale_alpha);
beta ~ normal(0.0, scale_beta);
sigma ~ exponential(loc_sigma);
// see Stan prior distribution suggestions
nu ~ gamma(2, 0.1);
// likelihood
y ~ student_t(nu, mu, sigma);
}
generated quantities {
// simulate data from the posterior
vector[N * use_y_rep] y_rep;
// log-likelihood posterior
vector[N * use_log_lik] log_lik;
for (i in 1:num_elements(y_rep)) {
y_rep[i] = student_t_rng(nu, mu[i], sigma);
}
for (i in 1:num_elements(log_lik)) {
log_lik[i] = student_t_lpdf(y[i] | nu, mu[i], sigma);
}
}
As noted in Heteroskedasticity, the Student-t distribution can be represented as a
scale-mixture of normal distributions, where the inverse-variances (precisions) follow
a Gamma distribution,
\[
\begin{aligned}[t]
y_i &\sim \dnorm\left(\mu_i, \omega^2 \lambda_i^2 \right) \\
\lambda^{-2} &\sim \dgamma\left(\nu / 2, \nu / 2\right)
\end{aligned}
\]
The scale mixture distribution of normal parameterization of the Student t distribution is useful for computational reasons.
A Stan model that implements this scale mixture of normal distribution representation of the Student-t distribution is lm_student_t_2.stan:
// lm_student_t_2.stan
// Linear Model with Student-t Errors
data {
// number of observations
int N;
// response
vector[N] y;
// number of columns in the design matrix X
int K;
// design matrix X
// should not include an intercept
matrix [N, K] X;
// priors on alpha
real scale_alpha;
vector[K] scale_beta;
real loc_sigma;
// keep responses
int use_y_rep;
int use_log_lik;
}
parameters {
// regression coefficient vector
real alpha;
vector[K] beta;
// regression scale
real sigma;
// 1 / lambda_i^2
vector[N] inv_lambda2;
// degrees of freedom;
// limit df = 2 so that there is a finite variance
real nu;
}
transformed parameters {
vector[N] mu;
vector[N] omega;
// observation variances
for (n in 1:N) {
omega[n] = sigma / sqrt(inv_lambda2[n]);
}
mu = alpha + X * beta;
}
model {
real half_nu;
// priors
alpha ~ normal(0.0, scale_alpha);
beta ~ normal(0.0, scale_beta);
sigma ~ exponential(loc_sigma);
nu ~ gamma(2, 0.1);
half_nu = 0.5 * nu;
inv_lambda2 ~ gamma(half_nu, half_nu);
// likelihood with obs specific scales
y ~ normal(mu, sigma);
}
generated quantities {
// simulate data from the posterior
vector[N * use_y_rep] y_rep;
// log-likelihood posterior
vector[N * use_log_lik] log_lik;
for (n in 1:num_elements(y_rep)) {
y_rep[n] = student_t_rng(nu, mu[n], omega[n]);
}
for (n in 1:num_elements(log_lik)) {
log_lik[n] = student_t_lpdf(y[n] | nu, mu[n], omega[n]);
}
}
Another reparameterization of these models that is useful computationally is The variance of the Student-t distribution is a function of the scale and the degree-of-freedom parameters. Suppose \(X \sim \dt(\nu, \mu, \sigma)\), then \[ \Var(X) = \frac{\nu}{\nu - 2} \sigma^2. \] So variance of data can be fit better by either increasing \(\nu\) or increasing the scale \(\sigma\). This will create posterior correlations between the parameters, and make it more difficult to sample the posterior distribution. We can reparameterize the model to make \(\sigma\) and \(\nu\) less correlated by multiplying the scale by the degrees of freedom. \[ \begin{aligned} y_i \sim \dt\left(\nu, \mu_i, \sigma \sqrt{\frac{\nu - 2}{\nu}} \right) \end{aligned} \] In this model, changing the value of \(\nu\) has no effect on the variance of \(y\), since \[ \Var(y_i) = \frac{\nu}{\nu - 2} \sigma^2 \frac{\nu - 2}{\nu} = \sigma^2 . \]
15.2.1 Examples
Estimate some examples with known outliers and compare to using a normal
See the data examples income_ineq, unionization, and econ_growth in the
associated jrnold.bayes.notes package.
data("econ_growth", package = "jrnold.bayes.notes")rec_union <-
recipe(union_density ~ left_government + labor_force_size + econ_conc,
data = unionization) %>%
step_center(everything()) %>%
step_scale(everything()) %>%
prep(retain = TRUE)union_data <- lst(
X = juice(rec_union, all_predictors(), composition = "matrix"),
y = drop(juice(rec_union, all_outcomes(), composition = "matrix")),
N = nrow(X),
K = ncol(X),
scale_alpha = 10,
scale_beta =rep(2.5, K),
loc_sigma = 1,
use_y_rep = 1,
use_log_lik = 1,
d = 4
)rec_econ_growth <-
recipe(econ_growth ~ labor_org + social_dem,
data = econ_growth) %>%
step_interact(~ labor_org * social_dem, sep = ":") %>%
step_center(everything()) %>%
step_scale(everything()) %>%
prep(retain = TRUE)econ_growth_data <- lst(
X = juice(rec_econ_growth, all_predictors(), composition = "matrix"),
y = drop(juice(rec_econ_growth, all_outcomes(), composition = "matrix")),
N = nrow(X),
K = ncol(X),
scale_alpha = 10,
scale_beta =rep(2.5, K),
loc_sigma = 1,
use_y_rep = 1,
use_log_lik = 1,
d = 4
)models <- list()
models[["lm_normal_1"]] <- stan_model("stan/lm_normal_1.stan")fits <- list()
fits[["econ_normal"]] <- sampling(models[["lm_normal_1"]], data = econ_growth_data)models[["lm_student_t_0"]] <- stan_model("stan/lm_student_t_0.stan")fits[["econ_t0"]] <- sampling(models[["lm_student_t_0"]], data = econ_growth_data, refresh = -1)models[["lm_student_t_1"]] <- stan_model("stan/lm_student_t_1.stan")
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:1:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Core:531:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:2:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/LU:47:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:3:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Cholesky:12:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Jacobi:29:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:3:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Cholesky:43:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/QR:17:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Householder:27:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:5:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/SVD:48:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:6:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Geometry:58:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core.hpp:14:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/core/matrix_vari.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat/fun/Eigen_NumTraits.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Dense:7:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Eigenvalues:58:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:12:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat.hpp:83:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/csr_extract_u.hpp:6:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Sparse:26:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/SparseCore:66:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:12:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat.hpp:83:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/csr_extract_u.hpp:6:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Sparse:27:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/OrderingMethods:71:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:12:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat.hpp:83:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/csr_extract_u.hpp:6:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Sparse:29:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/SparseCholesky:43:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:12:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat.hpp:83:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/csr_extract_u.hpp:6:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Sparse:32:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/SparseQR:35:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> In file included from file199a4ffb80c1.cpp:8:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/src/stan/model/model_header.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math.hpp:4:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/rev/mat.hpp:12:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat.hpp:83:
#> In file included from /Users/jrnold/Library/R/3.5/library/StanHeaders/include/stan/math/prim/mat/fun/csr_extract_u.hpp:6:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/Sparse:33:
#> In file included from /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/IterativeLinearSolvers:46:
#> /Users/jrnold/Library/R/3.5/library/RcppEigen/include/Eigen/src/Core/util/ReenableStupidWarnings.h:10:30: warning: pragma diagnostic pop could not pop, no matching push [-Wunknown-pragmas]
#> #pragma clang diagnostic pop
#> ^
#> 13 warnings generated.
#> ld: warning: directory not found for option '-L/usr/local/opt/llvm/lib/clang/5.0.0/lib/darwin/'fits[["mod_student_t_1"]]
#> NULL
fit_econ_t1 <- sampling(models[["lm_student_t_1"]], data = econ_growth_data, refresh = -1)models[["lm_student_t_2"]] <- stan_model("stan/lm_student_t_2.stan")fits[["econ_t2"]] <- sampling(models[["lm_student_t_2"]], data = econ_growth_data, refresh = -1)
#> Warning: There were 1 chains where the estimated Bayesian Fraction of Missing Information was low. See
#> http://mc-stan.org/misc/warnings.html#bfmi-low
#> Warning: Examine the pairs() plot to diagnose sampling problemscalc_loo <- function(x) {
ll <- extract_log_lik(x, "log_lik",
merge_chains = FALSE)
r_eff <- relative_eff(exp(ll))
loo(ll, r_eff = r_eff)
}
model_loo <- map(fits, calc_loo)
#> Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.
#> Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.
#> Warning: Some Pareto k diagnostic values are slightly high. See help('pareto-k-diagnostic') for details.
map(model_loo, ~ .x[["estimates"]]["elpd_loo", "Estimate"])
#> $econ_normal
#> [1] -22.7
#>
#> $econ_t0
#> [1] -22.8
#>
#> $econ_t2
#> [1] -22.2pars <- imap_dfr(fits, ~ mutate(tidyMCMC(.x, conf.int = TRUE), model = .y))
ggplot() +
geom_pointrange(data = filter(pars, str_detect(term, "^y_rep")) %>%
mutate(id = as.integer(str_extract(term, "\\d+"))),
mapping = aes(x = id, y = estimate, ymin = conf.low, ymax = conf.high, colour = model),
position = position_dodge(width = 0.2)) +
geom_point(data = tibble(y = econ_growth_data$y,
x = seq_along(y)),
mapping = aes(x = x, y = y)) +
coord_flip()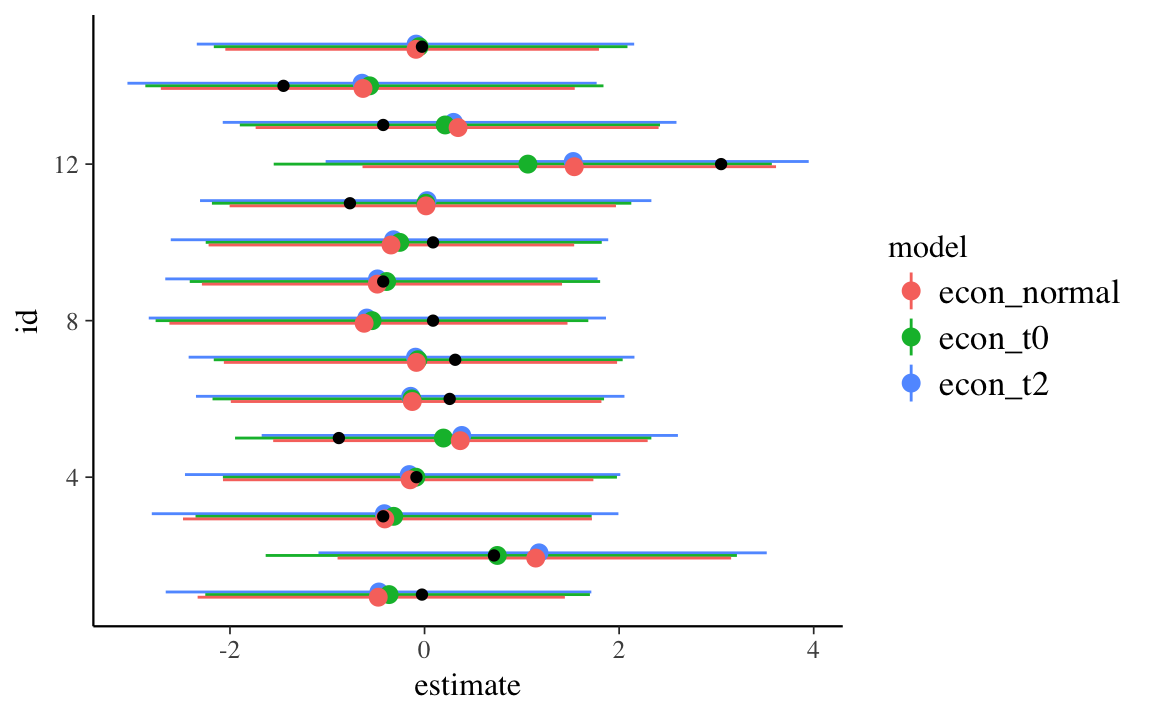
ggplot() +
geom_pointrange(data = filter(pars, str_detect(term, "^beta")),
mapping = aes(x = term, y = estimate, ymin = conf.low, ymax = conf.high, colour = model),
position = position_dodge(width = 0.2)) +
coord_flip()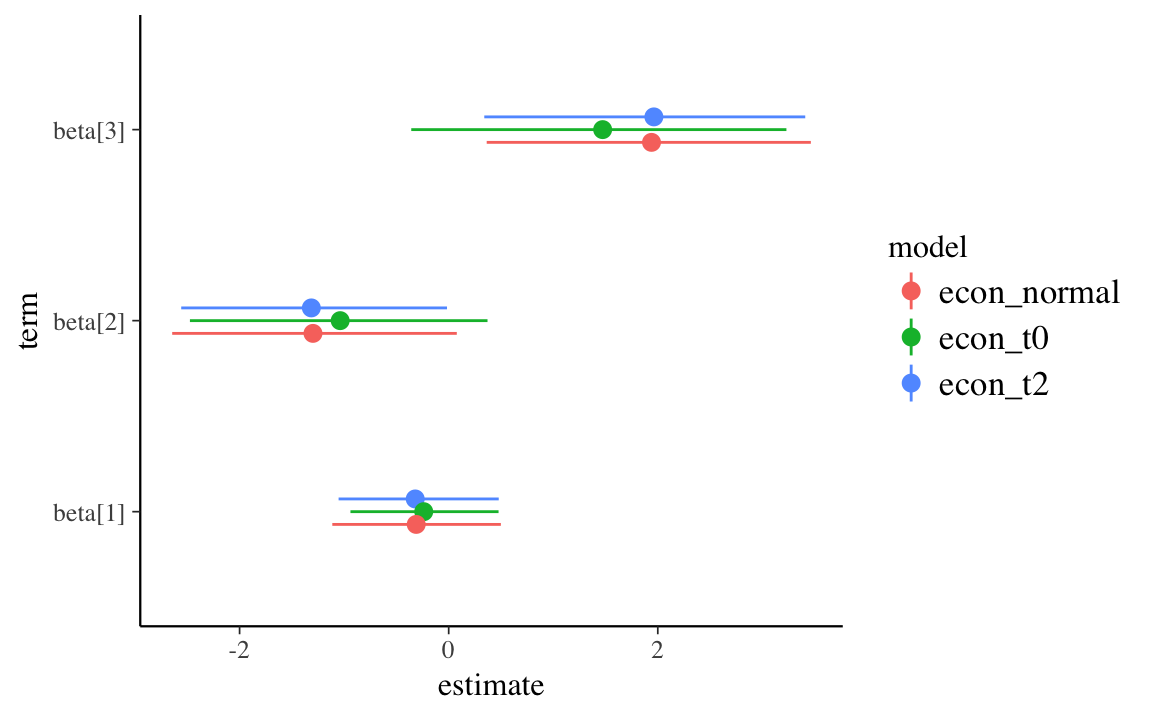
15.3 Robit
The “robit” is a “robust” bivariate model.(A. Gelman and Hill 2007, 125; Liu 2005) For the link-function the robit uses the CDF of the Student-t distribution with \(d\) degrees of freedom. \[ \begin{aligned}[t] y_i &\sim \dBinom \left(n_i, \pi_i \right) \\ \pi_i &= \int_{-\infty}^{\eta_i} \mathsf{StudentT}(x | \nu, 0, (\nu - 2)/ \nu) dx \\ \eta_i &= \alpha + X \beta \end{aligned} \] Since the variance of a random variable distributed Student-\(t\) is \(d / d - 2\), the scale fixes the variance of the distribution at 1. Fixing the variance of the Student-\(t\) distribution is not necessary if \(d\) is fixed, but is necessary if \(d\) were modeled as a parameter. Where \(\nu\) is given a low degrees of freedom \(\nu \in [3, 7]\), or a prior distribution.
15.4 Quantile regression
A different form of robust regression and one that often serves a different purpose is quantile regression.
Least absolute deviation (LAD) regression minimizes the following objective function, \[ \hat{\beta}_{LAD} = \arg \min_{\beta} \sum | y_i - \alpha - X \beta | . \] The Bayesian analog is the Laplace distribution, \[ \dlaplace(x | \mu, \sigma) = \frac{1}{2 \sigma} \left( - \frac{|x - \mu|}{\sigma} \right) . \] The Laplace distribution is analogous to least absolute deviations because the kernel of the distribution is \(|x - \mu|\), so minimizing the likelihood will also minimize the least absolute distances.
Thus, a linear regression with Laplace errors is analogous to a median regression. \[ \begin{aligned}[t] y_i &\sim \dlaplace\left( \alpha + X \beta, \sigma \right) \end{aligned} \] This can be generalized to other quantiles using the asymmetric Laplace distribution (Benoit and Poel 2017, @YuZhang2005a).
15.4.1 Questions
OLS is a model of the conditional mean \(E(y | x)\). A linear model with normal errors is a model of the outcomes \(p(y | x)\). How would you estimate the conditional mean, median, and quantile functions from the linear-normal model? What role would quantile regression play? Hint: See Benoit and Poel (2017 Sec. 3.4).
Implement the asymmetric Laplace distribution in Stan in two ways:
- Write a user function to calculate the log-PDF
- Implement it as a scale-mixture of normal distributions
15.5 References
For more on robust regression see A. Gelman and Hill (2007 sec 6.6), A. Gelman, Carlin, et al. (2013 ch 17), and Stan Development Team (2016 Sec 8.4).
For more on heteroskedasticity see A. Gelman, Carlin, et al. (2013 Sec. 14.7) for models with unequal variances and correlations. Stan Development Team (2016) discusses reparameterizing the Student t distribution as a mixture of gamma distributions in Stan.
The Double Exponential distribution still has a thinner tail than the Student-t at higher values.↩